Bruker has been hosting some fantastic webinars on getting to know their software and instruments during the COVID-19 shutdown (Found here). There are some fantastic little tips that come out of these webinars that to the average student (with only a few years of experience, and mostly with automated collection) may find confusing or duanting…This is not one of those tips.
Working with 2D Metabolomics data can be quite difficult, since the amount of data collected, processed, and stored can increase quite quickly. In the words of Clemens Anklin from Bruker during the tips and tricks webinar:
NMR Data is like a noble gas, it will expand to fill all available space
So how can we reduce the impact of our data on our storage? The problem is real, but the solution is imaginary. When you process your NMR data, especially NUS data, the file size jumps quite quickly because you are using the real and imaginary part of the data to process. However, once your processing is done, the imaginary serves little to no purpose for most users - so we can discard it.
To do this, open a file in Topspin from the directory you wish to condense. In this case, I have a folder which is for a completed project- perfect! Let’s see how we can cut down on storage:
Starting directory size: 7.33GB
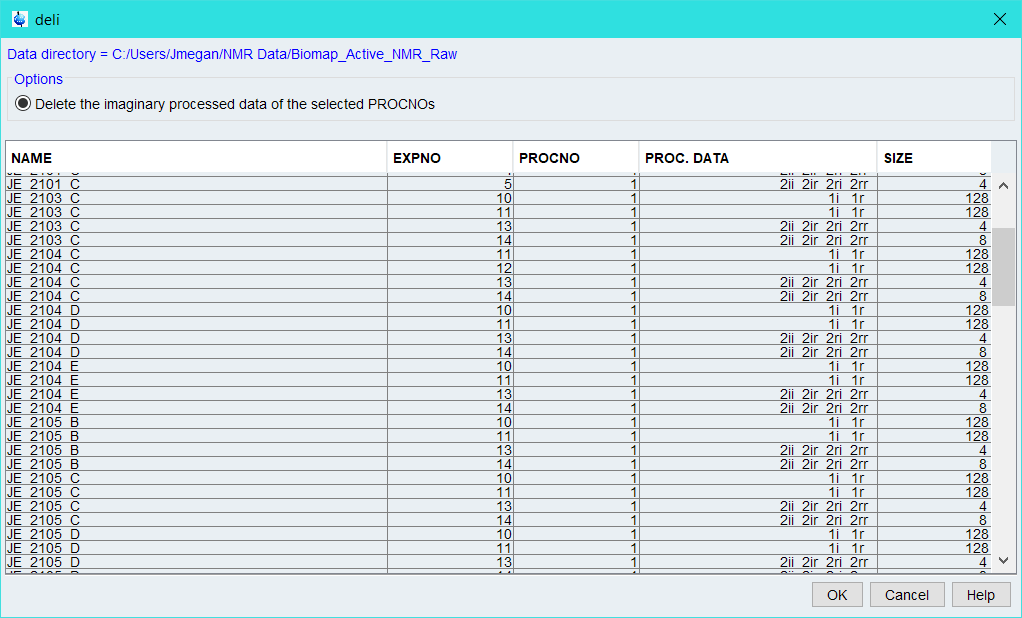
Opening a data file from my “Biomap Active” project, I then type “deli”. Although it sounds like I’m trying to have topspin reconstruct a sandwhich, it stands for delete imaginary. This then launches the following menu:
Since I don’t need any of this imaginary data at the moment, I select all the files and hit ‘ok’. There is no confirmation and it doesn’t take very long, but…
New directory size: 2.03GB.
If you ever need the imaginary part of the data back, you can simply re-calculate it by doing a hilbert transformation - type ‘xht1’ and/or ‘xht2’ to reconstruct it if you need it.